Researchers have recently made a significant advancement in imaging technology that is poised to revolutionize our understanding of the complexities of heart development and other biological processes. A new software tool, called clipping spline, provides unparalleled capabilities to view inside 3D images, enabling scientists to study intricate structures like never before. This dynamic and interactive tool allowed the research team to analyze previously unseen dynamics in the development of the embryonic mouse heart, using a specific type of 3D imaging technique known as optical coherence tomography (OCT).
The work, led by Shang Wang of the Stevens Institute of Technology, is crucial not only for understanding normal heart development but also for helping to devise new clinical strategies to address congenital heart diseases. These diseases, which are the most common type of birth defects, remain a significant health challenge worldwide. With new insights into how the heart develops, particularly during its early embryonic stages, researchers can more effectively manage congenital heart conditions and pave the way for innovative regenerative strategies. Such strategies may be key to repairing damaged heart tissue after events like heart attacks, contributing to improved cardiac function in affected patients.
The team’s work is detailed in a publication in Biomedical Optics Express, where they describe the clipping spline as an open-source, user-friendly software tool designed for visualizing complex 3D structures. Unlike traditional methods for visualizing 3D data, which typically offer static or difficult-to-navigate images, the clipping spline provides interactive, dynamic cutaway views of complex biological forms. This is particularly useful for imaging three-dimensional objects that curve or twist, such as the developing mouse heart, into a manageable single-cutaway view.
According to Andre Faubert, a research associate at Wang’s lab and a member of the team, 3D imaging is already a vital tool in the field of biomedicine. However, the team’s new approach opens up the potential to study not just 3D images, but 4D images—those that capture data over time. By incorporating the temporal aspect, researchers can monitor how biological structures develop, move, or evolve in real time. Though the clipping spline was initially demonstrated with 4D OCT data, its versatility allows it to be applied to volumetric images from a variety of imaging techniques, making it an asset for both biological research and clinical medical applications.
The need for such an innovative tool became clear when the team started analyzing data from OCT scans of the mouse heart at a critical stage known as cardiac looping. The cardiac looping phase marks a vital step in heart development, where the linear heart tube bends and twists into a complex, convoluted shape, setting the stage for the formation of heart chambers and blood flow. This stage is essential in understanding both normal heart development and various congenital defects that can result from disruptions during this phase.
Until now, studying the dynamics of the cardiac looping process has been challenging. Although this stage could be visualized using advanced imaging techniques, there were limited tools that could handle the complexity of these images in a meaningful way. The researchers’ new tool, the clipping spline, solves this problem by allowing the volume clipping of intricate 3D images. In simple terms, volume clipping is a computational method where parts of an image are selectively removed to expose structures inside, much like using a knife to cut into an object to see what’s inside.
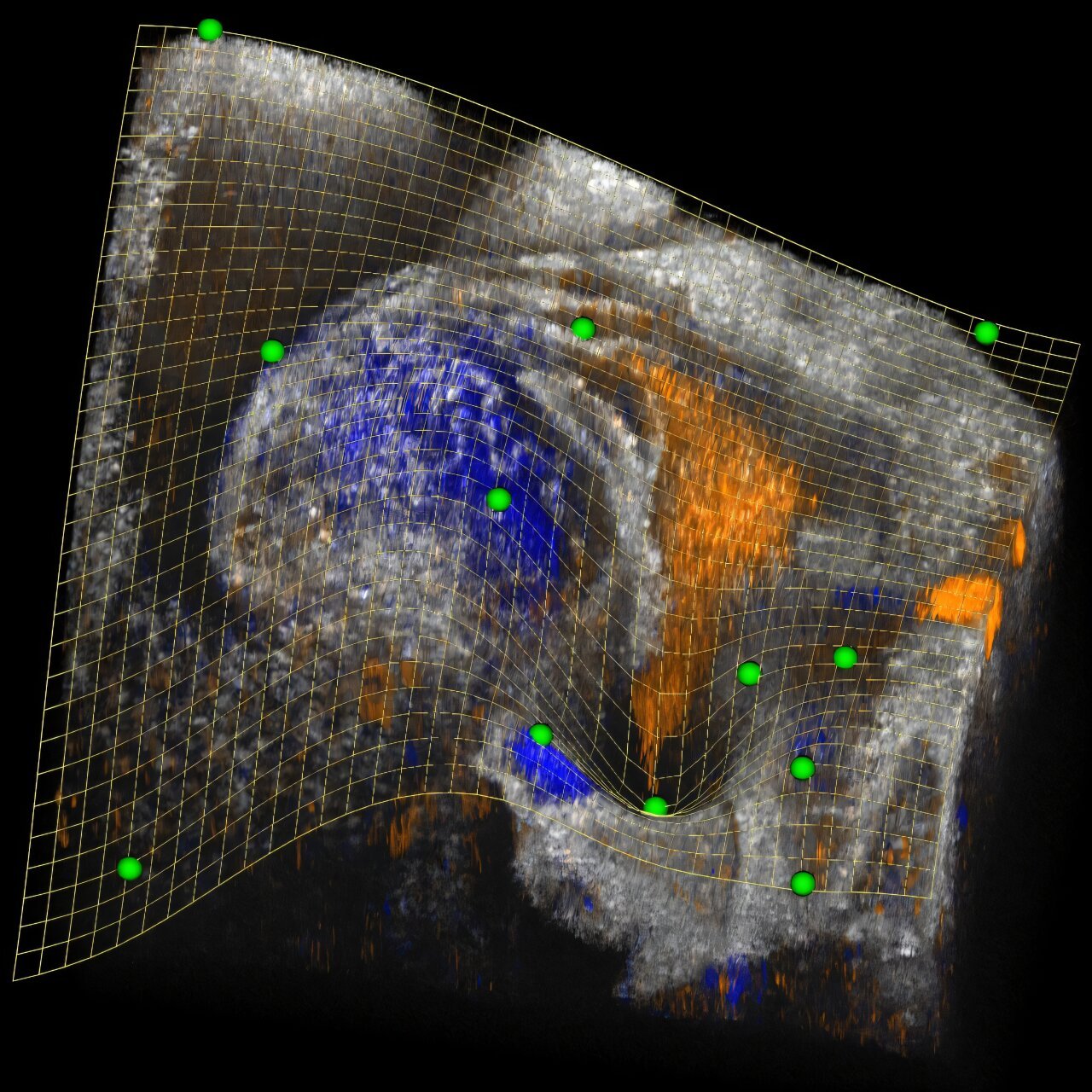
Performing volume clipping on complex biological structures is not a straightforward task, especially when it involves dynamic, twisting structures like the heart. Traditional clipping methods—usually relying on straight clipping planes—face limitations when trying to create clear cutaway views of irregular, non-planar forms. Clipping planes provide useful, but basic views by cutting a 3D object with a flat, geometrically simple boundary, which can’t represent curved or concave shapes well.
To address this, the team utilized a mathematical concept known as the thin plate spline (TPS) for the first time in the context of volume clipping. A TPS is a type of smooth 3D surface that connects a set of control points with minimal curvature. This smooth, flexible surface is not fixed; it can be adjusted interactively by modifying the positions or number of control points. TPS also enables complex geometric transformations, such as splitting, merging, or transitioning between different control points. As a result, the clipping spline offers a far more flexible and intuitive way of clipping complex 3D structures. Researchers can use this tool to dynamically refine and adjust cutaway views, enabling them to better visualize the heart’s internal features and study its development in detail.
In practice, the clipping spline can be used to follow the intricate movements of the embryonic mouse heart over time. The research team demonstrated its ability to track the heart’s evolution over 12.8 hours of development, using data collected from 712 time points. This allowed the researchers to observe, for the first time, how different parts of the convoluted heart tube developed and interacted with each other in 4D. In particular, the software helped visualize the formation of blood flow patterns within the heart, contributing to a deeper understanding of the mechanical aspects of heart development.
An exciting discovery made possible by this new tool was how the early heart’s inflow tracts converge to form the sinus venosus, a crucial structure that regulates blood flow into the developing heart. Being able to visualize and analyze these processes as they happen in real time could open new pathways for understanding both congenital heart defects and the regenerative processes of the heart.
“Watching these developmental processes unfold is truly amazing,” said Wang. “Not only does it provide deep insight into how the mammalian heart forms, but it also sparks new ideas and hypotheses, giving researchers a better chance to uncover fundamental principles of heart development.” Wang added that the understanding derived from studying the heart’s development can lead to advances in managing congenital conditions and contribute to critical research in other fields such as cancer biology and regenerative medicine.
The research team is confident that the clipping spline software will be a game-changer for the biomedical imaging community. By making it open-source, the team is allowing scientists from around the world to freely adopt and adapt the tool for their own research needs. Their next goal is to develop even more advanced image-processing methods to fully exploit the capabilities of the clipping spline. Furthermore, the team plans to continue applying the software to unravel the still-mysterious processes involved in embryonic heart development, ultimately working toward clinical breakthroughs that will improve the lives of countless individuals with heart conditions.
Reference: Andre C. Faubert et al, Clipping spline: interactive, dynamic 4D volume clipping and analysis based on thin plate spline, Biomedical Optics Express (2024). DOI: 10.1364/BOE.544231