In a groundbreaking study, a joint research team led by Sayuri Tsukahara and Tetsuji Kakutani of the University of Tokyo has uncovered crucial details about how retrotransposons—genetic elements that can “jump around” chromosomes and contribute to evolutionary changes—preferentially insert into the centromere of chromosomes. The research, published in the prestigious journal Nature, provides insight into the molecular mechanisms behind this phenomenon, expanding our understanding of how retrotransposons contribute to the evolution of eukaryotic genomes.
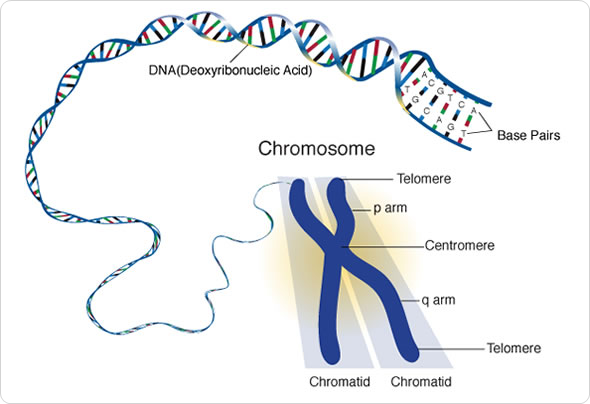
The centromere, a vital region of the chromosome, plays a crucial role during cell division. Structurally, it is the thin constricted part of the chromosome that separates its long and short arms, much like how the waist divides the upper and lower parts of the human body. The primary function of the centromere is to ensure accurate chromosome segregation during cell division. Remarkably, this role has been maintained across the eukaryotic lineage—the domain of organisms whose cells possess a membrane-bound nucleus, including plants, animals, and fungi.
While the function of the centromere has been conserved, its DNA sequence can exhibit substantial variations both within and across species, a phenomenon referred to as the “centromere paradox.” These variations have sparked ongoing intrigue among scientists, as they represent an area of genomes where significant differences can accumulate rapidly over evolutionary time. Retrotransposons, which can move or “jump” from one location in the genome to another, have long been identified as contributors to these variations, especially in the centromere. However, despite their importance, the precise mechanisms underlying retrotransposon insertions in the centromere have remained unclear until now.
To address this gap in understanding, the research team focused on the plant Arabidopsis lyrata, also known as the lyrate rockcress. This model plant species was chosen due to its well-characterized genomic features and recent advances in DNA sequencing technology that enabled detailed mapping of the centromere region, a resource that previously did not exist for many species.
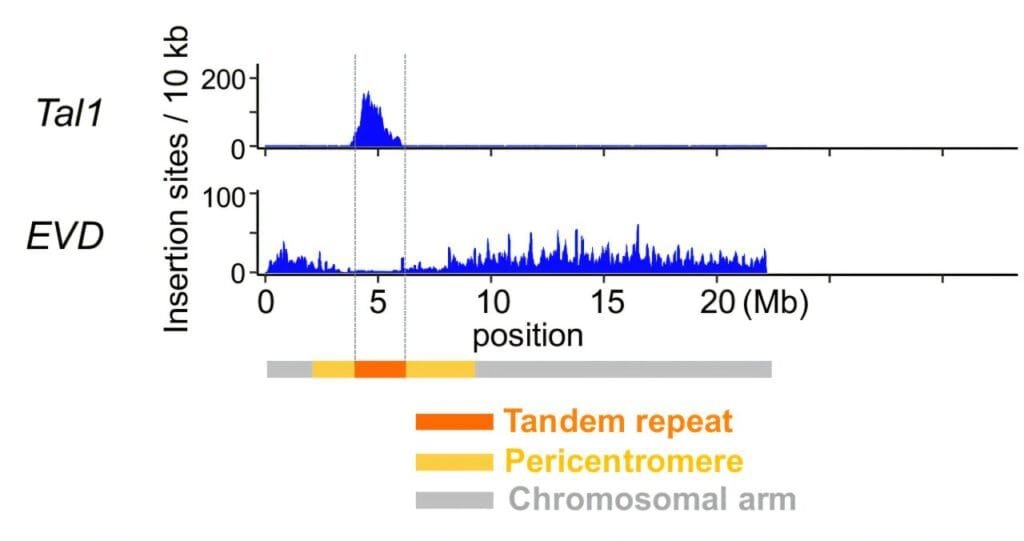
The research team’s investigation centered on two specific retrotransposons—Tal1 and EVD. These genetic elements were studied for their behavior in terms of where they integrate into the genome of Arabidopsis lyrata, particularly the centromere. Retrotransposons like Tal1 and EVD have long been known to make substantial contributions to genomic diversity and evolutionary processes. Their insertion into new locations in the genome can cause variations that may provide beneficial genetic diversity, facilitating evolutionary adaptation over time.
Through this study, the team employed a newly developed method known as TEd-seq, which allows for highly efficient detection of retrotransposon insertions within the genome. This technique, which had previously been used by some of the co-authors of the study, enabled the research team to map retrotransposon insertion sites with great precision. By utilizing reference data that had been recently assembled for the Arabidopsis lyrata centromere, they were able to more accurately track where retrotransposons like Tal1 and EVD integrated within the chromosome.
What the researchers discovered was highly revealing. Both Tal1 and EVD demonstrated significant integration biases, though their behavior differed in fascinating ways. Tal1 retrotransposons showed a strong preference for integrating directly into the centromere region of the chromosome, with almost no insertions in the chromosomal arm regions. On the other hand, EVD, although closely related to Tal1, showed a marked preference for integrating into the chromosomal arms rather than the centromere.
The findings suggest that the integration process for these two retrotransposons is not random but instead influenced by molecular factors that are distinct for each. This discovery was surprising because Tal1 and EVD are similar in structure, yet they exhibit differing insertion patterns in the chromosome. This strongly indicates that there are additional regulatory mechanisms at play that govern the specific targeting of retrotransposons to particular chromosomal regions, such as the centromere.
An even more intriguing result came when the researchers swapped a specific region—known as the C-terminal integrase region—between the two retrotransposons. In doing so, the integration patterns of Tal1 and EVD reversed. Tal1, which normally integrates into the centromere, began to prefer the chromosomal arm regions, while EVD preferentially inserted into the centromere, mirroring the behavior of Tal1. This experimental result suggests that the C-terminal integrase region plays a key role in determining where retrotransposons will insert within the chromosome.
These results highlight the sophisticated and intricate nature of retrotransposon integration mechanisms. The specific targeting of retrotransposons to the centromere region likely reflects a complex interaction between the molecular machinery responsible for retrotransposon mobility and the genomic architecture of the centromere. Understanding these mechanisms is crucial for gaining insights into how genomes evolve and how genetic elements influence this process over time.
As the research team continues to analyze the integration behaviors of retrotransposons, Tsukahara and Kakutani are enthusiastic about the next steps in their investigation. One of their main goals is to identify the factors that bind to retrotransposon Tal1 and facilitate its specific integration into the centromere. This could shed light on how centromeres, despite their variations across different species, maintain their functionality over evolutionary time. Additionally, they aim to explore whether retrotransposon insertions in the centromere are passed on preferentially to offspring, further unraveling the evolutionary impact of such genetic changes.
The implications of this research extend far beyond Arabidopsis lyrata and plant genomes. Retrotransposons are found in the genomes of a wide range of organisms, including humans, where they account for a significant portion of the DNA. Understanding the molecular mechanisms that govern retrotransposon integration could lead to broader insights into evolutionary biology, genome evolution, and the processes by which organisms adapt to changing environments. The study also underscores the importance of understanding the dynamics of transposable elements, which have long been recognized as potent drivers of genomic innovation and complexity.
Reference: Centrophilic retrotransposon integration via CENH3 chromatin in Arabidopsis, Nature (2024). DOI: 10.1038/s41586-024-08319-7