A team of scientists from multiple institutions has created a significant new resource designed to streamline the process of classifying patient tumor samples based on molecular features. This initiative, developed from the findings of The Cancer Genome Atlas (TCGA) Network, is a free and publicly accessible tool aimed at advancing cancer diagnosis and treatment. The resource provides classifier models capable of identifying cancer subtypes and holds the promise of accelerating the design of test kits specifically targeted for clinical trials and diagnostics. It represents a notable step forward because tumors that fall into different subtypes can respond quite differently to various cancer therapies, making accurate classification essential for optimizing treatment strategies.
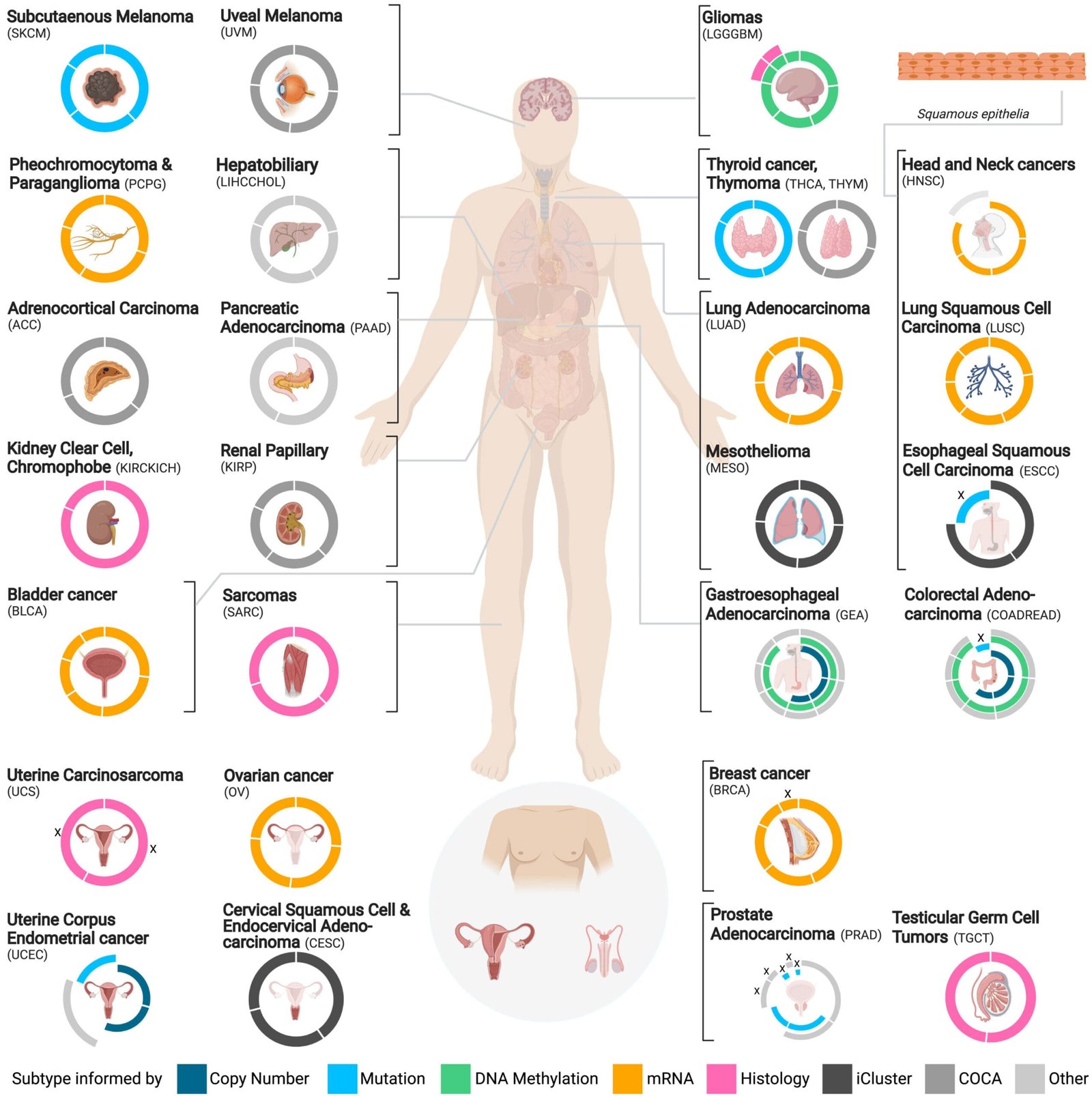
The new resource, which is the first of its kind, effectively bridges the gap between the wealth of data produced by TCGA and the practical application of that data in clinical settings. TCGA, an ambitious, National Cancer Institute-led project, sought to construct comprehensive molecular maps of 33 types of cancer. Traditional cancer classification systems focus primarily on the organ or tissue where the cancer originates. In contrast, TCGA utilized a multifaceted approach to cancer characterization, considering a broad range of genomic, epigenomic, proteomic, and transcriptomic factors to reveal more precise subtypes of cancer.
The study behind this development was recently published in the journal Cancer Cell, providing further insights into the creation of the tool and its potential applications. Dr. Peter W. Laird, a leading researcher at Van Andel Institute and the study’s lead corresponding author, highlighted the resource’s core objective: to provide the scientific and clinical communities with the tools they need to classify new cancer tumors according to the established molecular subtypes defined by TCGA. “With this resource, we aimed to provide the clinical and scientific communities with the tools to assign a newly diagnosed tumor to one of these established subtypes,” Dr. Laird said. “Our new resource will be a powerful asset for creating clinical assays based on the diverse molecular variations between cancers.” This availability of tools marks a step towards making these molecular subtype classifications clinically feasible and valuable.
Cancer subtyping through TCGA offers significant promise in cancer research. Traditionally, many treatment decisions were based on broad categories of cancer, but the recognition that cancers of similar organs might behave in vastly different ways depending on molecular underpinnings is revolutionary. Different cancer subtypes, even those within the same organ, can respond very differently to therapy, influencing patient survival rates, drug effectiveness, and side-effect profiles. As such, determining an individual cancer’s molecular profile can be critical to selecting the most effective therapeutic approach for that specific patient.
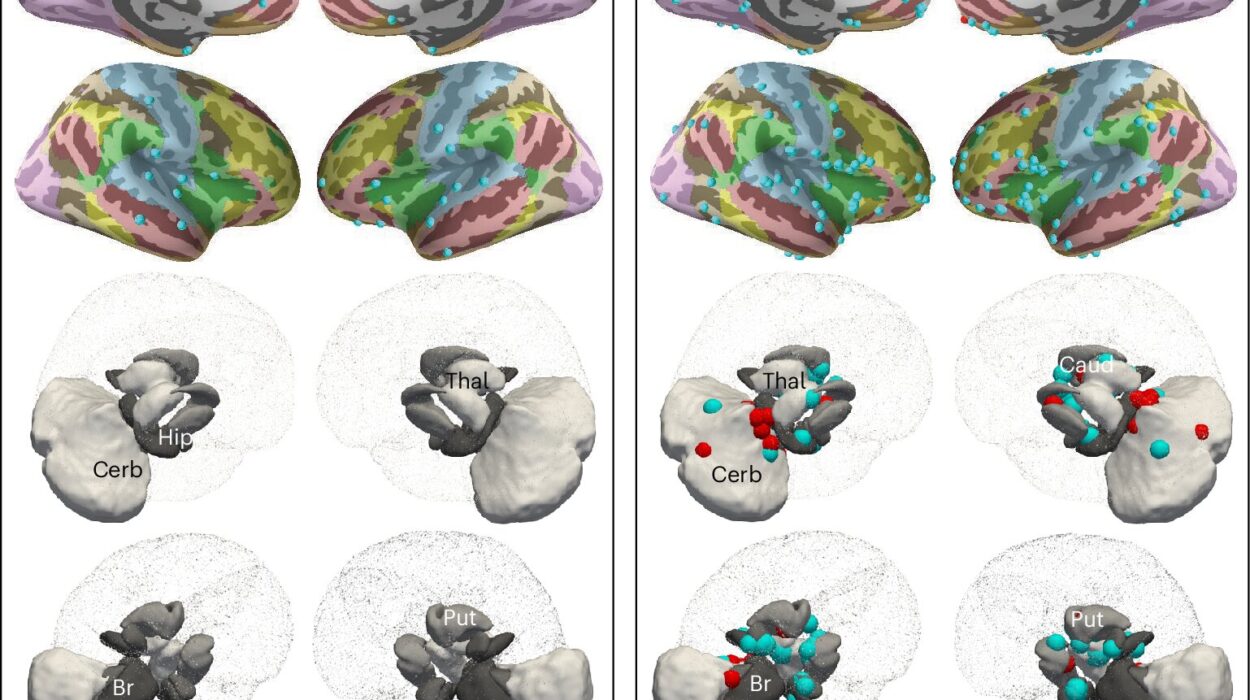
The resource leverages extensive data from 8,791 TCGA cancer samples representing 26 different cancer cohorts and encompassing 106 distinct cancer subtypes. By applying cutting-edge machine learning techniques, the team developed, tested, and curated models based on a vast number of features across several data categories—gene expression, DNA methylation, microRNA (miRNA) profiles, copy number variations, mutations, and multi-omics data. This deep data integration is pivotal because it provides a more holistic and detailed understanding of cancer biology, which is key to accurately subclassifying tumors. The resource includes a total of 737 high-performing classifier models, each corresponding to one of the cancer cohorts or subtypes. These models cover a variety of data types and training algorithms to ensure robustness and wide applicability.
The data in the resource will enable other scientists, physicians, and clinical practitioners to implement the classifiers on their own datasets, streamlining tumor classification processes. The ultimate goal of this innovation is not just to facilitate research but to provide clinicians with the tools they need to ensure better, more precise treatment. “A major element of this effort was working to ensure that these models could be deployed by other groups onto new datasets,” explained Dr. Kyle Ellrott, a collaborator from the Knight Cancer Institute at Oregon Health & Science University. “All too often this type of work is difficult to replicate or apply to new samples.” With this resource, clinical researchers now have access to a consistent, easy-to-use system for classifying cancer samples, making it more feasible to include molecular subtyping as a routine part of cancer treatment and research protocols.
This resource’s implications extend far beyond the academic world. As the healthcare community pushes for more personalized medicine, this tool could transform how doctors approach cancer diagnosis and treatment. Personalized or precision medicine aims to tailor treatment strategies based on an individual’s unique genetic makeup and the molecular features of their cancer. By using classifier models that associate molecular features with specific cancer subtypes, doctors can gain a clearer understanding of the likely behavior of a patient’s cancer, enabling them to recommend treatments most likely to yield positive outcomes. It also allows the design of better-targeted clinical trials, where researchers can identify patients whose tumors match specific subtypes and may respond more effectively to a particular drug.
The ability to map and subtype tumors based on extensive data—rather than relying on limited traditional methods—offers tremendous potential for improving early cancer detection, patient stratification for clinical trials, and the development of novel therapies tailored to specific cancer profiles. This tool could be used across a wide range of cancer types, making it an essential resource for any institution engaged in cancer research or patient care. Moreover, because the resource is freely available and publicly accessible, it holds the promise of democratizing cancer research and clinical applications worldwide. Researchers and clinicians, regardless of their location or resources, will be able to benefit from the robust, machine learning-enhanced models developed by this multidisciplinary team of scientists.
Reference: Classification of non-TCGA Cancer Samples to TCGA Molecular Subtypes Using Compact Feature Sets, Cancer Cell (2024). DOI: 10.1016/j.ccell.2024.12.002. www.cell.com/cancer-cell/fullt … 1535-6108(24)00477-X