Proteins, the molecular machines of life, are made up of chains of amino acids that fold into precise three-dimensional structures. These structures are essential for the proteins to perform their critical functions, such as catalyzing biochemical reactions, transporting molecules, repairing DNA, and supporting cellular structure. The process by which a protein folds is both intricate and essential to its function, but despite decades of research, it remains a complex puzzle.
Recent breakthroughs, however, are shedding new light on this process, providing insights into protein structure and function that could have significant implications for drug design, novel biomaterials, and a host of other applications. A study published in PRX Life, led by Corey O’Hern, a professor at Yale University, has made a key contribution to understanding protein folding by investigating the interior of globular proteins, offering new perspectives on how these structures form and how they might be manipulated for future technologies.
The Challenge of Protein Folding
Proteins are composed of sequences of amino acids, which fold into specific shapes to enable their function. However, the pathway from linear chain to fully folded protein is far from straightforward. The shape a protein adopts is critical—small changes in the protein’s structure can result in a loss of function or even disease. Researchers have long sought to understand how proteins fold into their unique, functional shapes and why this process is so precise.
One of the fundamental questions in protein folding is the arrangement of amino acids inside the protein, particularly within the core of globular proteins. These proteins, which are spherical or nearly spherical, are the most common type and are responsible for numerous biological processes. The question has always been: What determines the packing density of the amino acids in the protein core? Why do proteins seem to settle into a specific packing fraction, and why is this fraction so consistent across different proteins?
New Insights from Computational Models
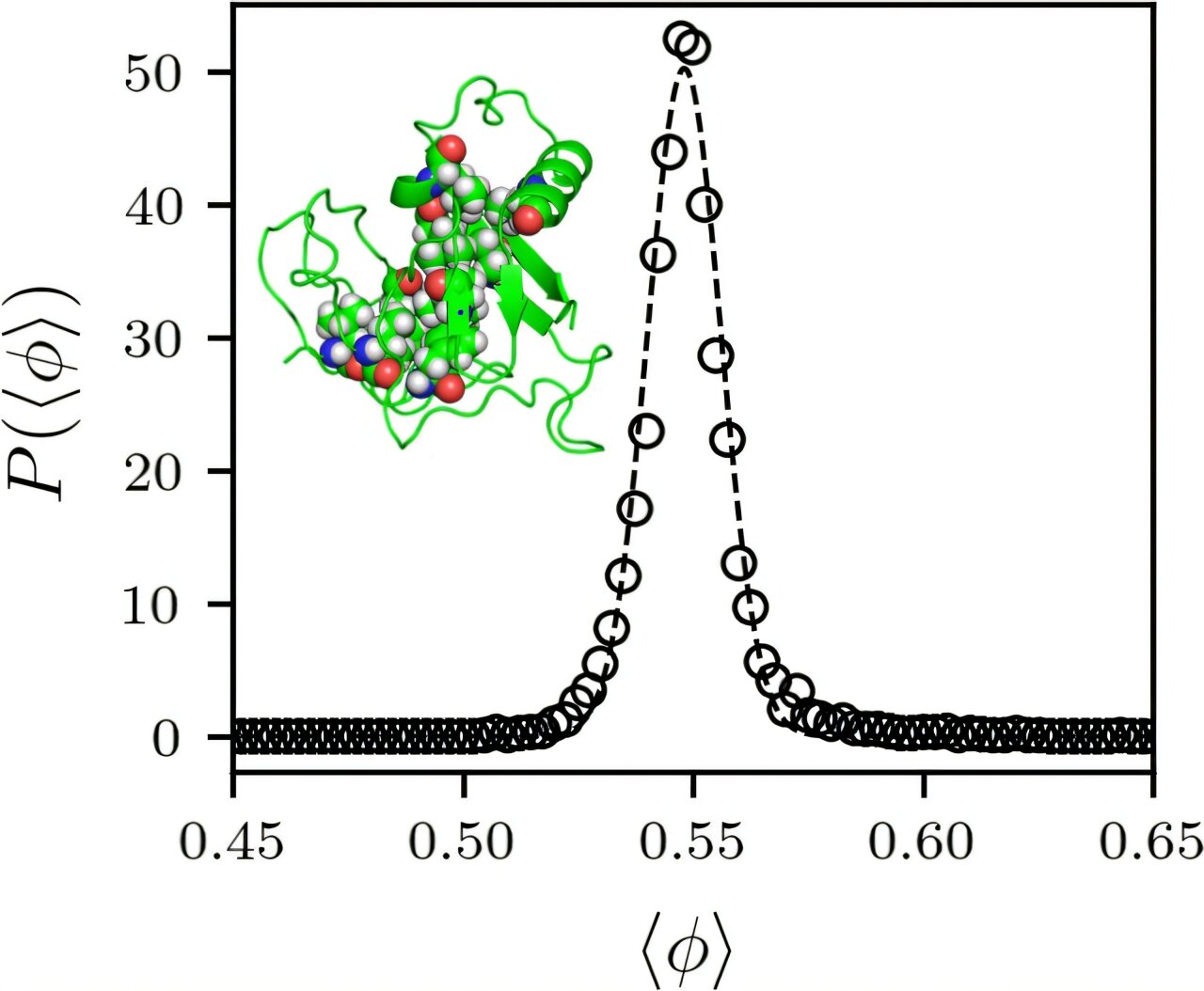
In an effort to answer these questions, the research team led by O’Hern developed computational models to analyze all globular proteins stored in the Protein Data Bank (PDB)—a comprehensive online resource of protein structures. These models allowed the team to assess the packing density of the protein cores, particularly focusing on how tightly the amino acids are packed into the interior.
What they found was striking: regardless of the specific protein, the core packing fraction was consistently about 55%. This means that in a given protein, 55% of the space within the protein core is occupied by atoms, and the remaining 45% is empty space. This seemingly universal value for the packing fraction raised two important questions: Why is it so consistent across different proteins, and why is it specifically 55%?
The Physics of Protein Folding and Core Packing
The researchers’ answer to this puzzle lies in the physical principles governing protein structure. According to O’Hern, the 55% packing fraction results from a phenomenon known as “jamming.” In simple terms, jamming occurs when particles (in this case, the amino acids in a protein) cannot be packed any more tightly together due to the physical constraints of their shapes.
Unlike spherical objects that can pack into tight, dense arrangements (such as the well-known close-packing of spheres at around 64%), the amino acids that make up proteins have more complex shapes. Some amino acids are roughly spherical, but the majority are elongated and have rough surfaces due to the bonded hydrogen atoms on their side chains. The result is that these irregularly shaped amino acids cannot pack as efficiently as spherical particles, leading to a lower packing fraction.
The team’s analysis showed that the jamming of these amino acids—the point at which they cannot be packed any more tightly—is what leads to the consistent 55% packing fraction observed in the protein cores. This explains why proteins, despite their vast diversity in function and structure, tend to follow this same packing pattern.
Exploring the Possibility of More Efficient Packing
While the 55% packing fraction is a consistent feature of proteins under normal physiological conditions, the study also suggests that there could be ways to push beyond this limit. The research team explored the possibility of packing proteins more densely by altering the conditions under which proteins fold.
For example, studies have shown that proteins subjected to high pressures—such as those found in deep ocean hydrothermal vents—can increase their core packing fraction to 58-60%. This increased packing density could play a role in understanding the origins of life, as these conditions are thought to mimic those of the early Earth, where the first organic molecules may have formed.
Alex Grigas, a Ph.D. candidate in O’Hern’s lab and lead author of the study, pointed out that while the packing fraction of proteins has generally been assumed to be capped at 55% under normal conditions, it’s possible that with the right modifications to the solvent, pressure, or temperature, proteins could be engineered to achieve higher packing densities. This opens the door to new methods for designing proteins with enhanced properties or novel functions.
Implications for Protein Design and Therapeutic Applications
This research has broad implications for the field of protein engineering and therapeutic design. Currently, protein design often focuses on creating novel sequences of amino acids to construct proteins with specific functions. However, O’Hern’s findings suggest that researchers could also manipulate the folding conditions themselves—such as solvent environments, pressure, and temperature—to create new protein structures and enhance their functions.
This opens up exciting possibilities for drug design and biomaterials, as proteins with optimized core packing could have enhanced stability, functionality, and resistance to degradation. In the future, scientists might be able to design proteins that perform more efficiently in specific conditions, such as in extreme environments, or that exhibit novel biological activities.
Additionally, this research could help scientists design better therapeutic proteins, such as enzymes or antibodies, that could be used in treating diseases. For example, proteins with more efficient packing could potentially be more stable and easier to produce in large quantities for medical applications.
The Road Ahead
While this study provides important insights into protein folding and core packing, much remains to be explored. Future research will likely focus on understanding how these findings can be translated into practical applications in protein design. This could involve testing different folding conditions in the lab to see how proteins with different packing fractions perform in real-world scenarios, particularly in the context of therapeutic applications and biomaterials.
Moreover, scientists will continue to explore the potential of manipulating the physical environment in which proteins fold, allowing for the creation of proteins with customized structures and functions that could be used in everything from drug delivery systems to the development of sustainable materials.
In conclusion, this research represents a significant step forward in our understanding of protein folding. By revealing the physics behind the packing density of protein cores and offering new avenues for manipulating this process, the study opens up a wealth of possibilities for the future of protein design, drug development, and materials science. The ability to engineer proteins with new and enhanced properties could revolutionize medicine and biotechnology in the years to come.
Reference: Alex T. Grigas et al, Protein Folding as a Jamming Transition, PRX Life (2025). DOI: 10.1103/PRXLife.3.013018