The complexity and biodiversity of microbial life are vast, yet still largely underexplored, particularly in terms of genomic diversity. Researchers from the U.S. Department of Energy’s Joint Genome Institute (JGI), located at Berkeley Lab, have conducted a comprehensive study assessing the current state of microbial genomic diversity, providing crucial insights into what we’ve learned over the past 30 years of sequencing, and offering a roadmap for further exploration. The findings, published in Science Advances, indicate that much of the diversity within bacteria and archaea has yet to be fully represented in genomic databases and calls for a renewed focus on experimental microbiology.
A Snapshot of Microbial Genomic Diversity
Microorganisms, including bacteria and archaea, play essential roles in various global cycles, from nutrient cycling to influencing climate dynamics. These organisms, despite being invisible to the naked eye, are fundamental to maintaining ecosystem stability and supporting human innovation, particularly in fields like agriculture, biofuels, and medicine. Despite their critical roles, the vast majority of microbial life remains undercharacterized.
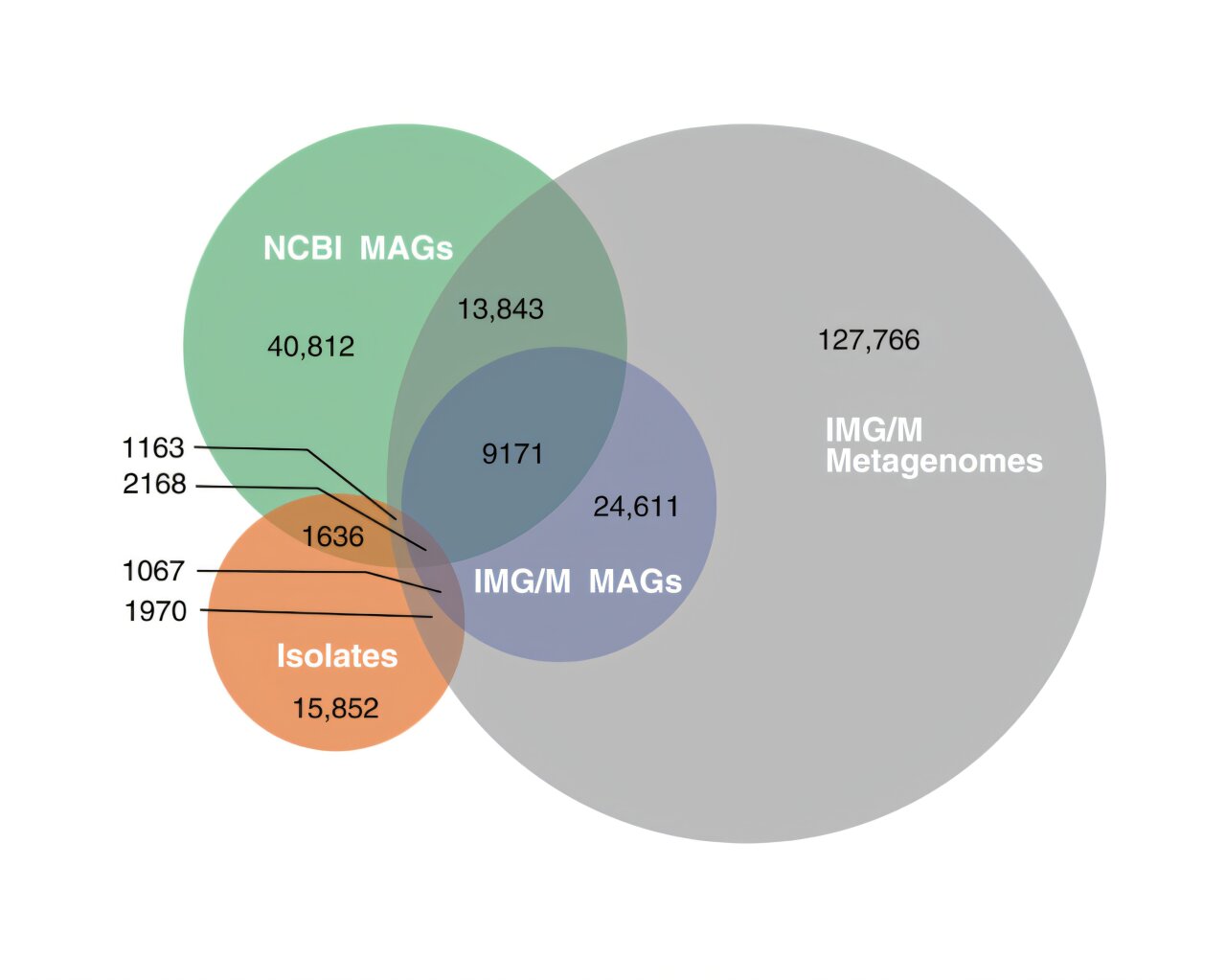
The study conducted by the JGI research team, led by Dongying Wu, Rekha Seshadri, Nikos Kyrpides, and Natalia Ivanova, evaluated more than 1.8 million bacterial and archaeal genomes compiled over three decades of sequencing efforts. Their goal was to assess the extent to which the microbial diversity currently represented in genomic databases provides a comprehensive snapshot of all possible microbial species. The team’s analysis utilized publicly available genomic data stored in databases such as GenBank (maintained by the National Center for Biotechnology Information, NCBI) and JGI’s own Integrated Microbial Genomes & Microbiomes (IMG/M) platform.
Using 5 conserved, protein-coding marker genes, the team analyzed the bacterial and archaeal datasets and found an interesting gap in the available genomic diversity. While progress has been made, Wu and Seshadri both remarked that despite three decades of sequencing, the available datasets represent only a small portion of the total microbial diversity, leaving significant gaps in our knowledge.
The Findings: A “Deep Dive” Into the Microbial Genomes
Through an extensive analysis of more than 1.8 million genomes, including both isolate genomes and metagenome-assembled genomes (MAGs), the researchers made several important observations. Isolate genomes, representing organisms grown in laboratory conditions, accounted for just a fraction of the actual microbial diversity.
For bacteria, only 9.73% of the predicted diversity was represented in isolate genomes. In contrast, MAGs, which are computationally derived from environmental DNA and offer a glimpse of genomes that cannot yet be cultivated in the lab, represented about 49% of the estimated bacterial diversity. This means nearly half of the bacterial diversity could potentially be recovered through further analysis of environmental samples. However, more than 40% of bacterial diversity remains without genomic representation in public databases, underscoring how much is still left to explore and document.
For archaea, the diversity was similarly underrepresented, with isolate genomes only capturing 6.55% of the estimated diversity. MAGs did somewhat better, representing about 57% of the archaeal diversity. However, roughly 36% of archaeal diversity remains absent from genomic databases.
The comparison of the two groups highlighted the substantial gaps in our microbial genomic knowledge. This reinforces the idea that microbial biodiversity is more expansive than we currently realize, and the known datasets likely represent only a tiny fraction of the microbial world.
Why the Microbial World Remains Underexplored
The reasons for these gaps in microbial genomic data are multifaceted. First, it’s important to understand that not all microorganisms can be easily cultured. Many species of bacteria and archaea live in environments where replicating the conditions for growth in a laboratory setting is challenging or even impossible. For example, some microorganisms thrive at extreme temperatures, pressures, or salinities, conditions that are difficult to mimic in a typical lab environment. As such, many microbial species, particularly those that exist in complex natural environments like soil, oceans, and the human gut, have eluded cultivation and remain invisible to traditional genomic approaches.
Advancements in next-generation sequencing technologies over the last several decades have improved our ability to capture microbial genomes directly from environmental samples, without the need for cultivation. Metagenomic approaches, which rely on extracting DNA from entire microbial communities, have significantly expanded our knowledge by enabling the sequencing of thousands of microbial genomes at once. However, the genomes obtained via metagenomics, called metagenome-assembled genomes (MAGs), are often incomplete or fragmented, and may not provide an exact match to a specific, cultivated strain.
Despite these challenges, MAGs have been crucial in expanding the catalog of known microbial genomes. According to the researchers, these genomes have led to major advances in microbial genomics by bridging the gap between uncultivable species and genomic data. Nevertheless, the MAGs are still computationally derived and lack the hands-on, experimental validation that comes with cultivating the species themselves.
The Road Ahead: Improving Our Understanding of the Microbial World
The implications of these findings go beyond basic scientific curiosity. Understanding microbial diversity is foundational to improving various industries, particularly bioengineering, where microbes are used for biofuels and bioproducts. In medicine, expanding our knowledge of the microbial world can lead to better probiotics, treatments for diseases, and more effective ways to manipulate microbial communities for health benefits.
Yet for this to be achieved, there is a clear need to move beyond computational studies and toward more experimental and hands-on microbiological research. The team’s study emphasizes that while MAGs have vastly expanded our understanding of microbial diversity, there remains an urgent need for targeted explorations to cultivate more species in the lab. Without the effort to actually cultivate microbial species, their potential applications cannot be fully realized.
The authors argue that microbial culturing must be reinvigorated. A combination of computational techniques for identifying microbial species of interest and more laboratory work for their experimental validation is critical. By identifying areas of the microbial world that have yet to be adequately studied, scientists could use new methods to cultivate previously elusive species.
Seshadri highlights the importance of a “treasure map” approach. “We’ve drawn out the treasure map,” she said. This means that researchers can now point directly to environmental samples that have the highest likelihood of yielding unknown microbial species and reinvest resources into the experimental cultivation of these organisms. This strategy could dramatically improve the speed and breadth with which we uncover the full depth of microbial diversity.
The Importance of Environmental Exploration
While there is significant focus on improving sequencing technology, Ivanova pointed out that much of the unexplored microbial diversity lies in specific environments—places where we may not have yet collected sufficient data. By directing future environmental sampling efforts to these “treasure spots,” researchers can enhance the collection of new samples and increase their chances of recovering previously unknown species for further study.
Overall, the JGI team’s study highlights the complexity of microbial life and the vast unknown that still lies ahead. While tremendous progress has been made, our understanding of the microbial world is still in its early stages. To realize the full potential of this microbial treasure trove, a concerted effort combining computational genomics, environmental exploration, and traditional experimental microbiology is essential.
Conclusion
The diversity of microbial life, although vastly cataloged through modern sequencing technologies, remains far from fully understood. Despite decades of advancements, a large proportion of bacterial and archaeal species have not yet been represented in genomic databases, particularly among organisms that have not been cultivated. While MAGs and computational approaches have played a transformative role in uncovering microbial diversity, there is still much work to be done. To bridge the gaps in our knowledge, researchers advocate for a revival of hands-on microbiology combined with computational discovery. Only through a coordinated, multidisciplinary effort will we unlock the full potential of the microbial world and its applications across medicine, agriculture, bioengineering, and beyond.
Reference: Dongying Wu et al, A metagenomic perspective on the microbial prokaryotic genome census, Science Advances (2025). DOI: 10.1126/sciadv.adq2166