Over the past two decades, large-scale genetic studies have made tremendous strides in identifying genetic variants linked to a wide array of human traits and diseases. These efforts have resulted in the identification of tens of thousands of DNA variants associated with thousands of diseases, allowing researchers to develop a deeper understanding of the genetic factors underlying human health and disease. However, despite these advances, applying this genetic knowledge to develop targeted therapies has remained a significant challenge. In particular, the ability to modify or correct these genetic variants in a precise and efficient manner has been limited by the lack of effective molecular tools that can target specific genetic alterations without causing unintended consequences.
A breakthrough in overcoming these challenges came from researchers at the Broad Institute of MIT and Harvard, Massachusetts General Hospital, and Harvard University. This team of scientists took an innovative approach to tackling the problem of targeting genetic variants associated with diseases. They focused on building a highly diverse and comprehensive library of molecular compounds that could be mined for potential treatments that target disease-related genetic variants in novel ways. Thanks to advancements in chemistry, they were able to create a library of over 3 million compounds. These compounds were specifically designed to bring two proteins together, using one protein as a “shield” to stabilize the other, with the aim of reversing the disease-causing effects associated with certain genetic variants.
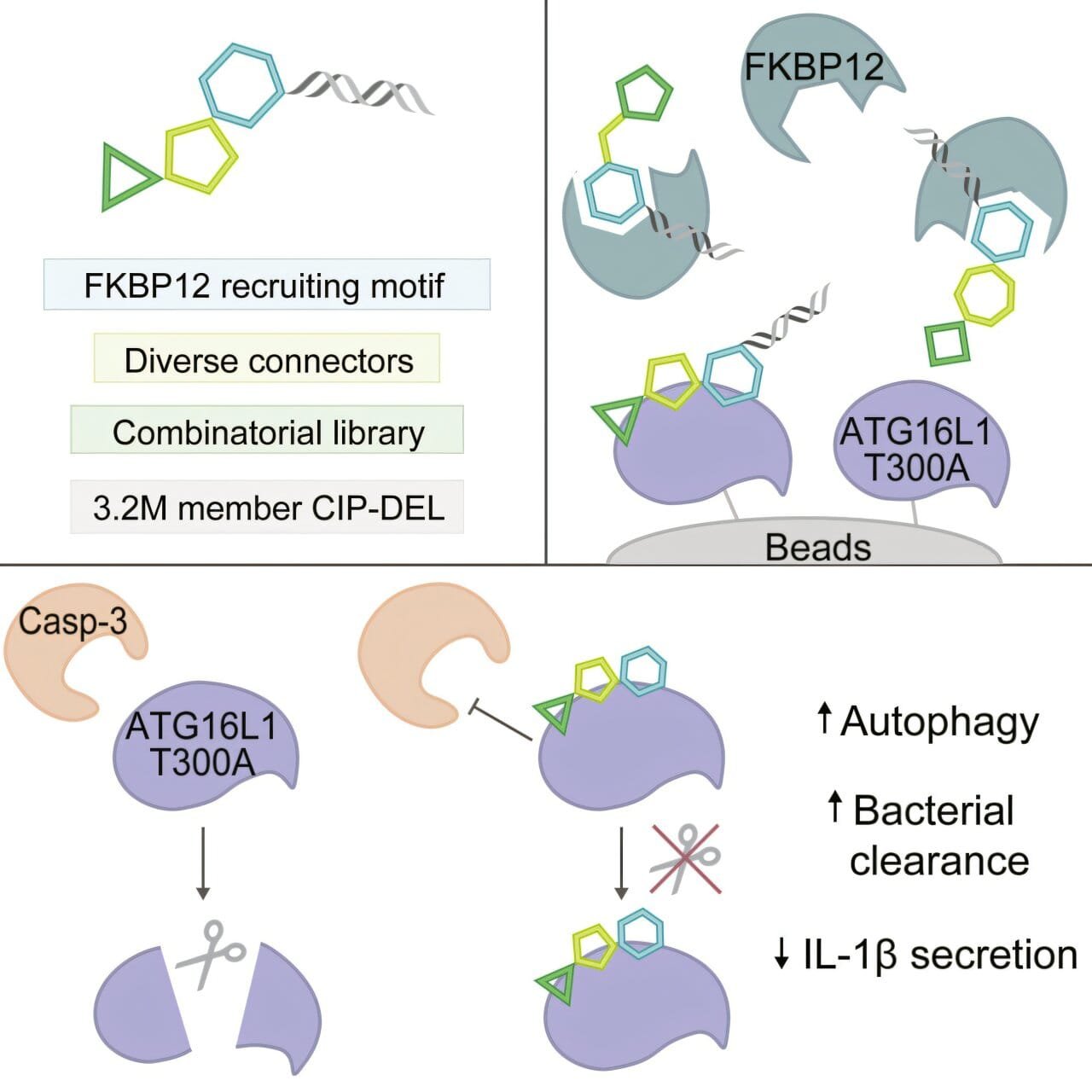
One of the key aspects of the team’s approach was their use of a class of compounds known as chemical inducers of proximity (CIPs). These include molecular glues and bifunctional compounds, which work by bringing together two proteins within the cell—proteins that would not normally interact with one another. The primary goal is to use these compounds to target disease-associated genetic variants, either by stabilizing or modifying the activity of the altered proteins, and potentially providing therapeutic benefits for a variety of genetic disorders.
Traditionally, CIP libraries have focused on using these compounds to target dysfunctional proteins and facilitate their degradation through the cell’s waste disposal systems, a mechanism known as the proteasome-mediated degradation. However, in this study, the researchers sought to go beyond simply degrading the target proteins. Instead, they designed their library to focus on compounds that would stabilize rather than degrade disease-related proteins, restoring their normal function and potentially offering a more effective therapeutic strategy.
The team chose FKBP12, a well-known protein that is abundant in human cells, as the “presenter” protein for their experiment. When a CIP compound engages FKBP12, the researchers hypothesized that it could act as a shield, protecting the target protein from enzymatic breakdown. By stabilizing the target protein in this way, the compound could help reverse the harmful effects of certain genetic variants without causing side effects associated with protein degradation. This represents a promising strategy for treating diseases caused by protein dysfunction, and the compound library developed by the team was designed to explore a wide range of potential interactions between the compounds and various target proteins.
To further enhance the specificity and potential therapeutic value of their approach, the team also utilized DNA barcoding techniques to track the compounds in large experimental screens. DNA-encoded libraries allow for the efficient screening of millions or even billions of compounds, enabling the identification of those that have the potential to target specific disease-related variants. By attaching a unique DNA barcode to each compound in the library, the researchers were able to identify those that were most likely to interact with the target proteins. They also incorporated a variety of linkers, choosing rigid rather than flexible chains to connect the chemical components of each compound, which allowed for greater control over the compounds’ binding interactions.
One of the most significant applications of this approach involved the study of a genetic variant associated with Crohn’s disease, a chronic inflammatory condition that primarily affects the gastrointestinal tract. The specific variant of interest, ATG16L1 T300A, was already known to impair a process called autophagy—an essential cellular mechanism that removes waste and harmful bacteria from cells. Prior research in the laboratory of Ramnik Xavier, a professor at Harvard Medical School and Massachusetts General Hospital, had shown that this particular variant of ATG16L1 makes the protein more susceptible to cleavage by caspase-3, an enzyme involved in the regulation of various cellular processes. The result is impaired autophagy, leading to cellular dysfunction and contributing to the development of Crohn’s disease.
In their screen of the CIP library, the researchers identified a compound that could interact specifically with the ATG16L1 T300A variant. Further tests in human and mouse cells demonstrated that the compound was able to stabilize the protein, protecting it from caspase-3 cleavage. As a result, the compound successfully reversed the impaired autophagy associated with this genetic variant, restoring cellular processes to normal function without impacting other important functions of caspase-3.
This result offers exciting potential for treating diseases that are caused by genetic variants that impair protein stability and function. The strategy developed by the research team could be extended to target a wide variety of genetic variants linked to other diseases, making it a powerful tool in the search for new treatments. The use of molecular glues and CIPs provides a more precise method for targeting disease-associated variants, with the possibility of directly modifying or stabilizing disease-related proteins rather than relying on degradation-based strategies.
As the study progresses, the researchers plan to further test their compound in animal models, to explore its therapeutic potential and evaluate its ability to provide effective treatments for Crohn’s disease. They also hope that their work will inspire other researchers to expand the library and customize it for different diseases. By selecting presenter proteins that are only present in specific cell types or tissues, it may be possible to refine these compounds so that they target only disease-affected areas of the body, offering a much more targeted and precise form of therapy.
Looking ahead, this work has the potential to usher in a new era of drug development that takes advantage of the wealth of genetic information available and combines it with cutting-edge molecular chemistry. The approach of using molecular glues and DNA-encoded libraries has the potential to revolutionize the treatment of genetic diseases, allowing for more effective, precise, and personalized therapies. This collaborative effort, which blends chemical biology with human genetics and disease understanding, represents the next step forward in the quest to cure genetically related diseases and treat them at their molecular source.
Ultimately, by continuing to innovate and explore new molecular approaches like CIPs, researchers could gain the ability to tackle a wide range of diseases with unprecedented accuracy and precision. The molecular toolbox they have developed holds tremendous promise for both researchers and patients alike, offering new hope for more effective therapies and, potentially, new ways of treating previously intractable genetic diseases. As the team continues to refine and expand their compound library, the possibilities for this new class of therapeutic interventions remain vast and promising, opening new avenues for the treatment of human disease.
Reference: Zher Yin Tan et al, Development of an FKBP12-recruiting chemical-induced proximity DNA-encoded library and its application to discover an autophagy potentiator, Cell Chemical Biology (2025). DOI: 10.1016/j.chembiol.2024.12.002