Researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius-Maximilians-Universität (JMU) in Würzburg have made groundbreaking discoveries regarding how gut bacteria regulate sugar metabolism, providing a more profound understanding of the intricate dynamics within the human intestinal microbiota. Their focus was on Bacteroides thetaiotaomicron, a common and pivotal species of gut bacteria, which plays a significant role in breaking down complex carbohydrates, or polysaccharides. The study identifies a critical protein, RbpB, and a group of small ribonucleic acids (sRNAs) collectively known as FopS, that regulate metabolic processes essential for these bacteria’s adaptation to changing nutrient availability in the gut. These findings, recently published in Nature Communications, open up exciting avenues for developing therapeutic strategies to harness the microbiota for improved health.
The human gut, often referred to as the “second brain,” is not only responsible for digestion but also plays an integral role in immune system regulation, nutrient absorption, and overall well-being. Central to its functioning is the gut microbiota—a dynamic community of trillions of microorganisms, including bacteria, viruses, and fungi. Among these, bacteria like Bacteroides thetaiotaomicron stand out for their ability to metabolize complex dietary fibers into simpler compounds, which can be absorbed by the human body or utilized by other microorganisms within the gut ecosystem.
However, the gut environment is far from static. Nutrient availability fluctuates due to dietary changes, fasting periods, or even microbial competition. For gut bacteria to thrive and maintain their metabolic contributions, they must adjust their biochemical processes in response to these fluctuations. While significant research has explored the transcriptional mechanisms governing how gut bacteria adapt, the post-transcriptional regulation of these adaptations has remained largely elusive.
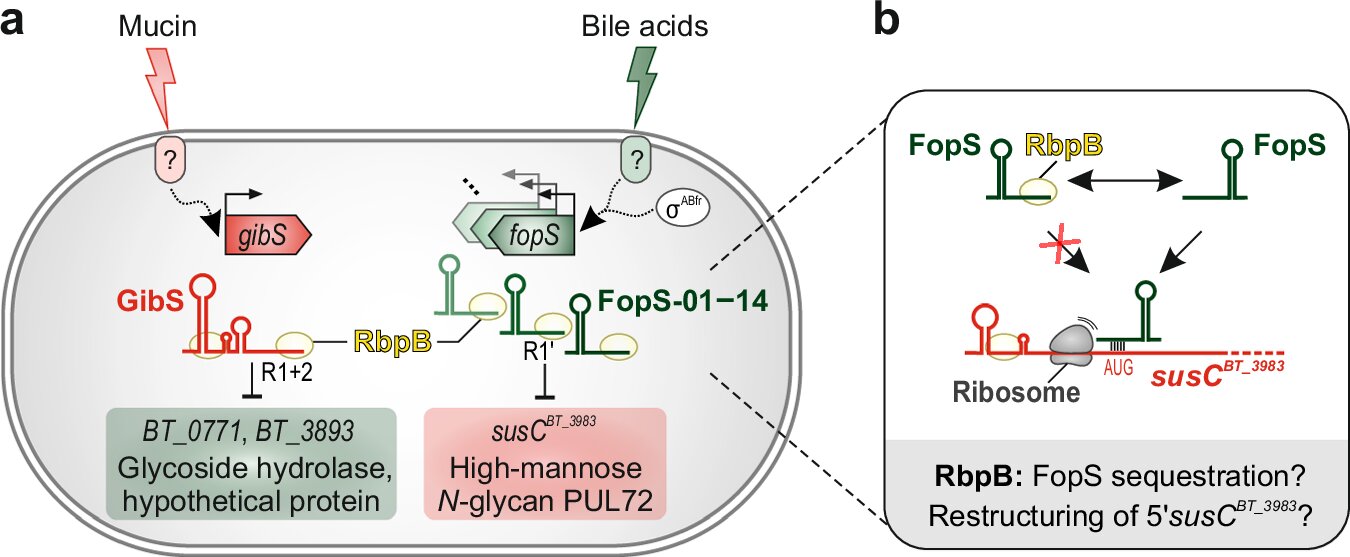
In their study, the researchers at HIRI and JMU tackled this knowledge gap, focusing on the regulatory mechanisms within Bacteroides thetaiotaomicron. This bacterium is uniquely equipped to bind, degrade, and import specific polysaccharides thanks to a set of highly specialized multiprotein complexes encoded in its genome at specific regions called polysaccharide utilization loci (PULs). These PUL complexes are critical for the bacterium’s ability to colonize and persist within the gut. While previous research emphasized transcriptional control of these PUL genes, the team delved into how these genes are regulated after transcription, uncovering a sophisticated RNA-based network.
Central to their findings was the identification of the RNA-binding protein RbpB. Experiments demonstrated that RbpB is indispensable for B. thetaiotaomicron’s survival and effective colonization of the gut. The protein interacts with numerous cellular transcripts, orchestrating their function and ensuring the bacteria can respond effectively to nutritional changes. This regulatory influence is further augmented by the involvement of 14 related non-coding RNA molecules, collectively known as FopS (family of paralogous sRNAs). Together, RbpB and FopS regulate key metabolic pathways, ensuring optimal adaptation to fluctuating nutrient supplies.
The absence of RbpB was found to severely impair B. thetaiotaomicron’s ability to colonize the gut, underlining its central role in microbial fitness. By controlling the degradation and utilization of complex sugars, this RNA-protein interplay not only facilitates bacterial survival but also contributes to the overall stability of the gut ecosystem. For instance, the breakdown products of polysaccharides serve as essential nutrients for both the host and other microbiota members, showcasing the interdependence of various species within the microbiome.
The researchers employed a combination of in vitro and in vivo experiments, working alongside collaborators from Vanderbilt University and the University of Toronto, to unravel the intricate details of this RNA-regulated metabolic control system. Their analysis highlights the complexity of bacterial adaptation strategies, which extend beyond transcriptional control into the realm of RNA-based mechanisms. This discovery not only enriches our understanding of microbial gene regulation but also establishes a new framework for studying the broader functional capabilities of gut bacteria.
Looking forward, the team plans to delve deeper into the structural biology of RbpB to better understand its RNA-binding properties and interaction networks. Additionally, they aim to investigate the potential parallels between RbpB and similar RNA-binding proteins in other dominant gut microbiota species. By uncovering these central post-transcriptional hubs, researchers hope to gain insights into the general principles of microbial adaptation and their role in maintaining gut health.
The implications of this research are profound. With better knowledge of how gut bacteria regulate their metabolism, scientists could design targeted therapeutic interventions to modulate microbiota activity. For example, understanding the RNA-based mechanisms of B. thetaiotaomicron could inspire new approaches to treat or prevent conditions such as inflammatory bowel disease (IBD), obesity, and metabolic disorders, which are often linked to microbiota imbalances. Moreover, manipulating bacterial gene and protein functions might pave the way for enhancing nutrient absorption or developing microbiota-based therapies tailored to individual dietary needs.
The findings also underscore the potential for leveraging the gut microbiota in combating infectious diseases. By understanding the competitive dynamics of commensal and pathogenic bacteria, researchers could develop strategies that favor the beneficial microbes, effectively using them as a biological shield against harmful invaders.
Reference: Ann-Sophie Rüttiger et al, The global RNA-binding protein RbpB is a regulator of polysaccharide utilization in Bacteroides thetaiotaomicron, Nature Communications (2025). DOI: 10.1038/s41467-024-55383-8